
An imbalance of charge within the polymer causes electrostatic repulsion between neighboring chromatin regions that promote interactions with positively charged proteins, molecules, and cations. The positively charged histone cores only partially counteract the negative charge of the DNA phosphate backbone resulting in a negative net charge of the overall structure.
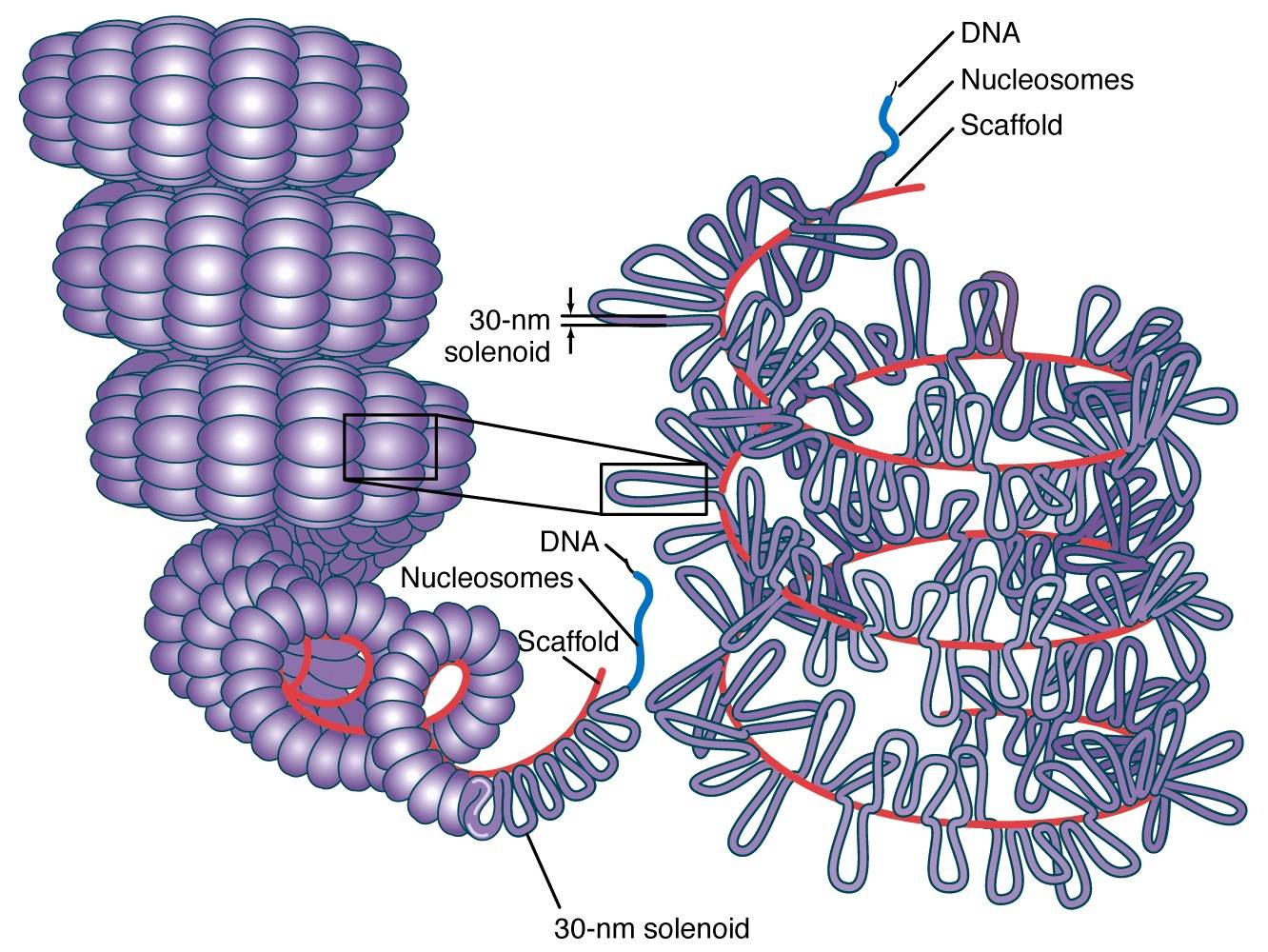
Most modifications occur on histone tails. Histone proteins are the basic packers and arrangers of chromatin and can be modified by various post-translational modifications to alter chromatin packing ( histone modification). There is limited understanding of chromatin structure and it is active area of research in molecular biology.ĭynamic chromatin structure and hierarchy Basic units of chromatin structure the structure of chromatin within a chromosomeĬhromatin undergoes various structural changes during a cell cycle. Epigenetic modification of the structural proteins in chromatin via methylation and acetylation also alters local chromatin structure and therefore gene expression. Regions of DNA containing genes which are actively transcribed ("turned on") are less tightly compacted and closely associated with RNA polymerases in a structure known as euchromatin, while regions containing inactive genes ("turned off") are generally more condensed and associated with structural proteins in heterochromatin. The local structure of chromatin during interphase depends on the specific genes present in the DNA. During interphase, the chromatin is structurally loose to allow access to RNA and DNA polymerases that transcribe and replicate the DNA. The overall structure of the chromatin network further depends on the stage of the cell cycle. Prokaryotic cells have entirely different structures for organizing their DNA (the prokaryotic chromosome equivalent is called a genophore and is localized within the nucleoid region). For example, spermatozoa and avian red blood cells have more tightly packed chromatin than most eukaryotic cells, and trypanosomatid protozoa do not condense their chromatin into visible chromosomes at all. Many organisms, however, do not follow this organization scheme.
#Scaffold protein h1 update
Please help update this article to reflect recent events or newly available information. This article's factual accuracy may be compromised due to out-of-date information.


 0 kommentar(er)
0 kommentar(er)
